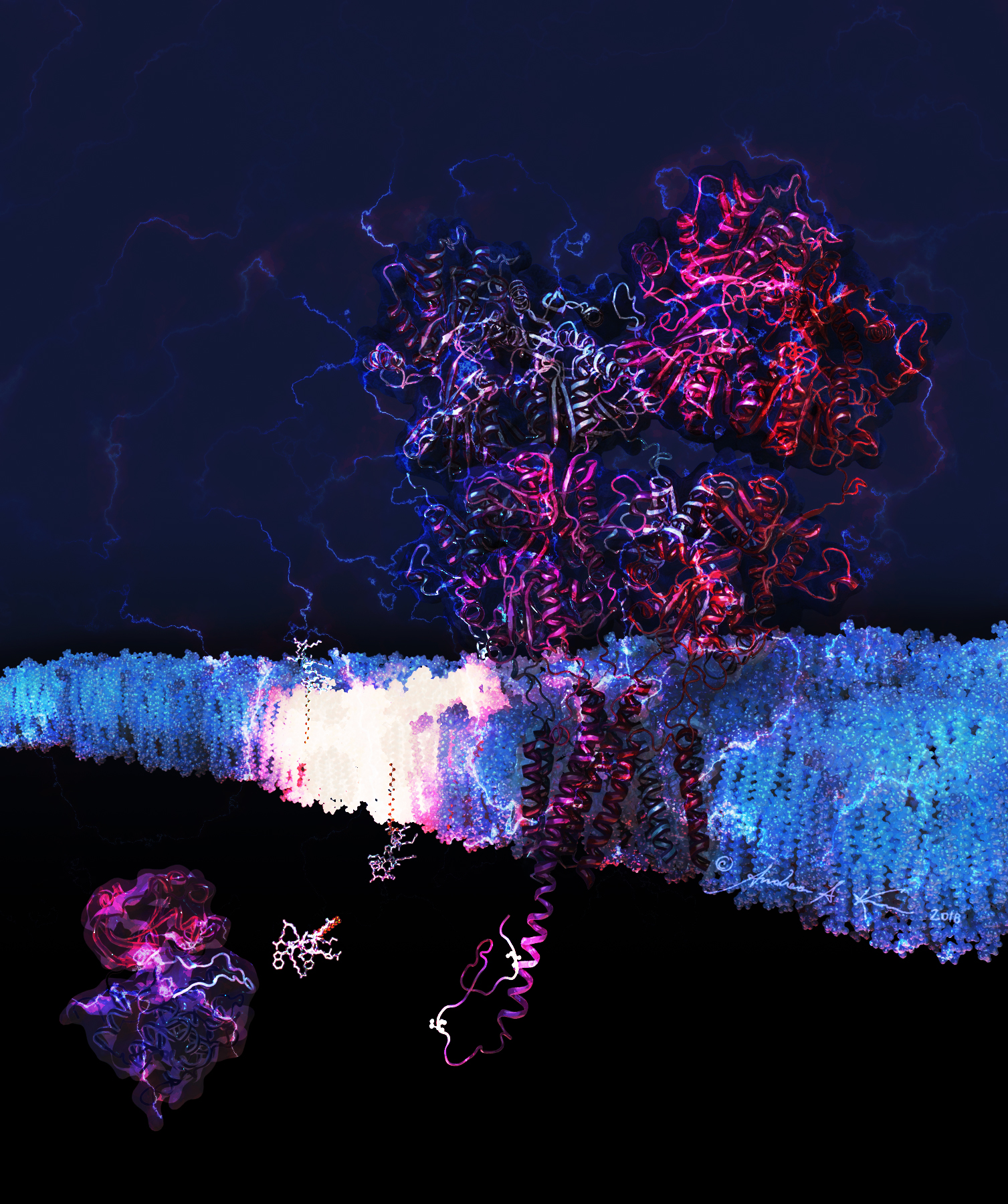
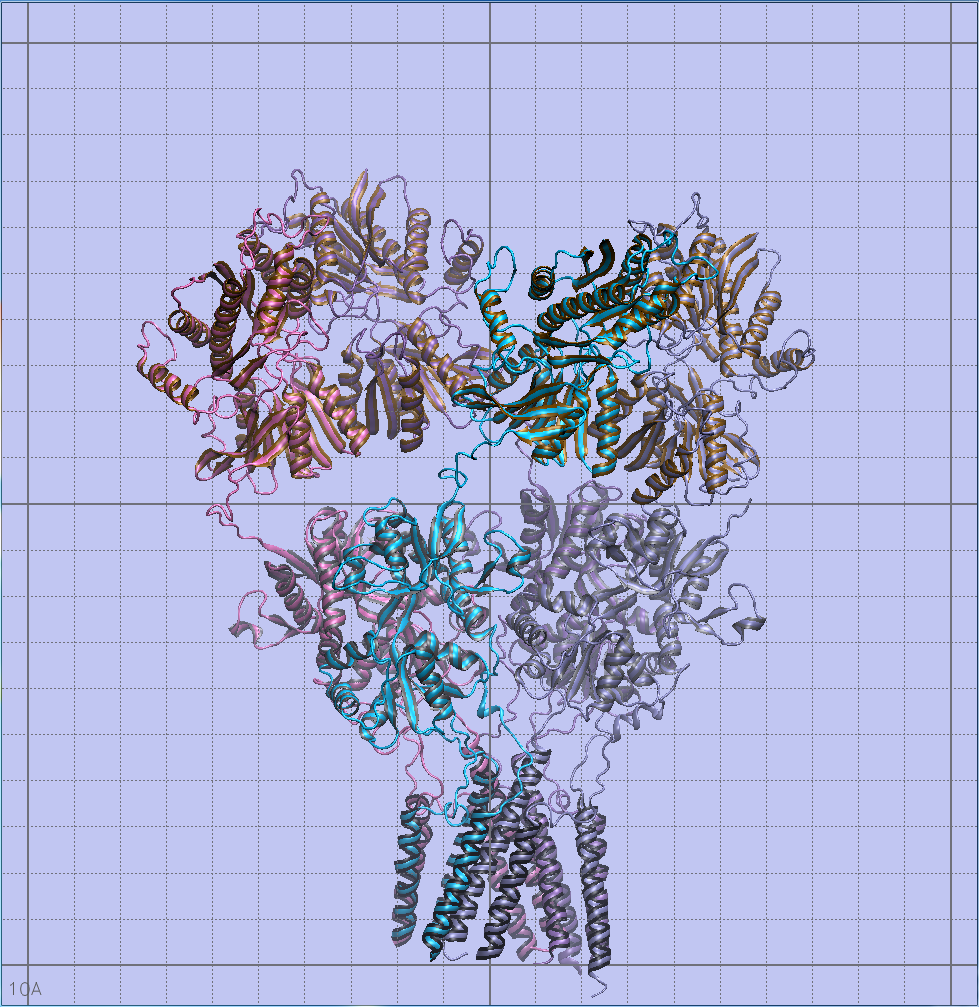Sketches and preliminary research
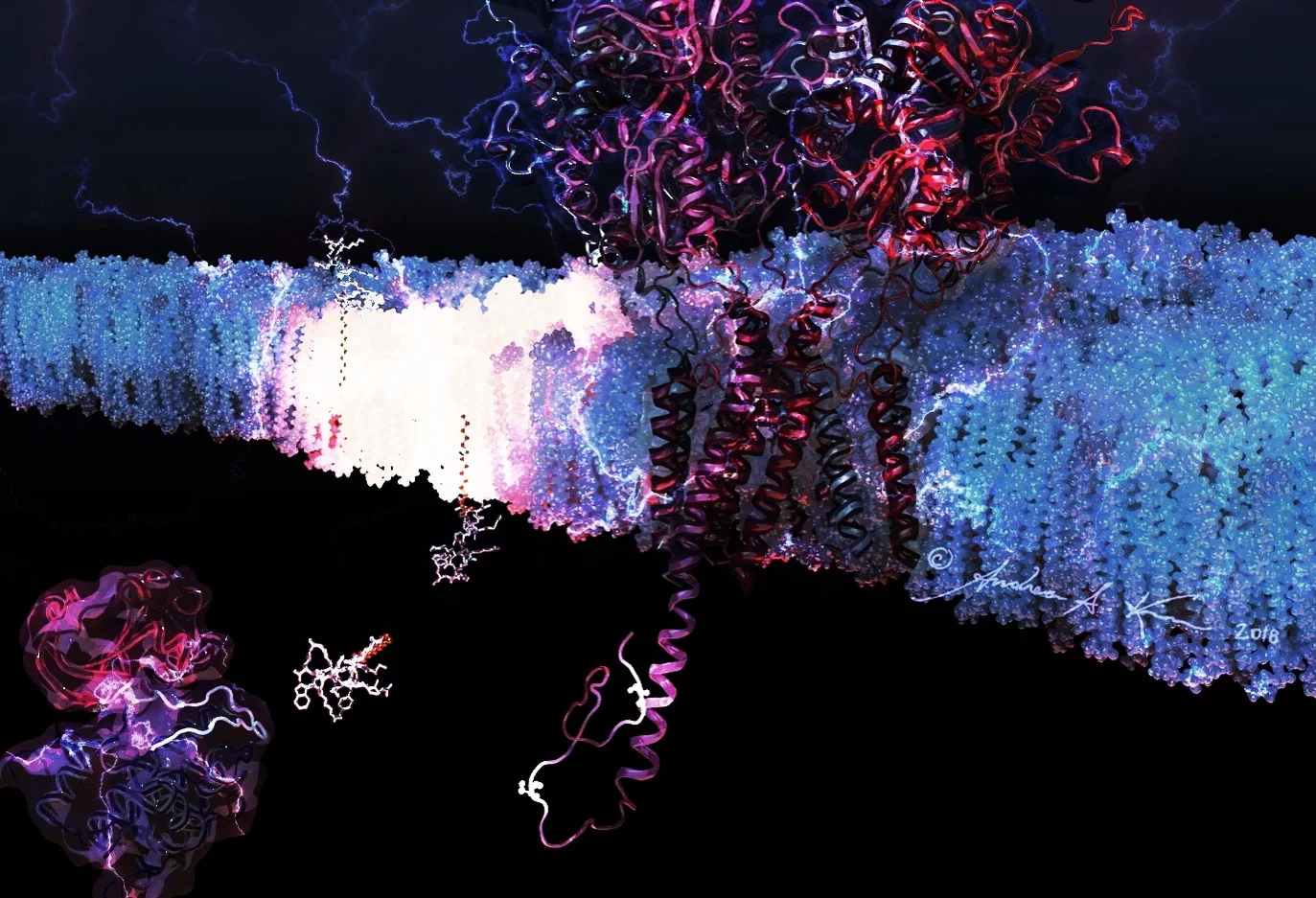
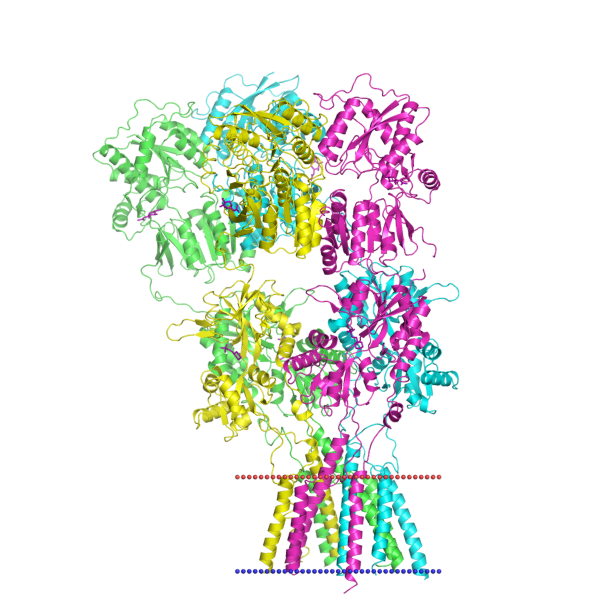
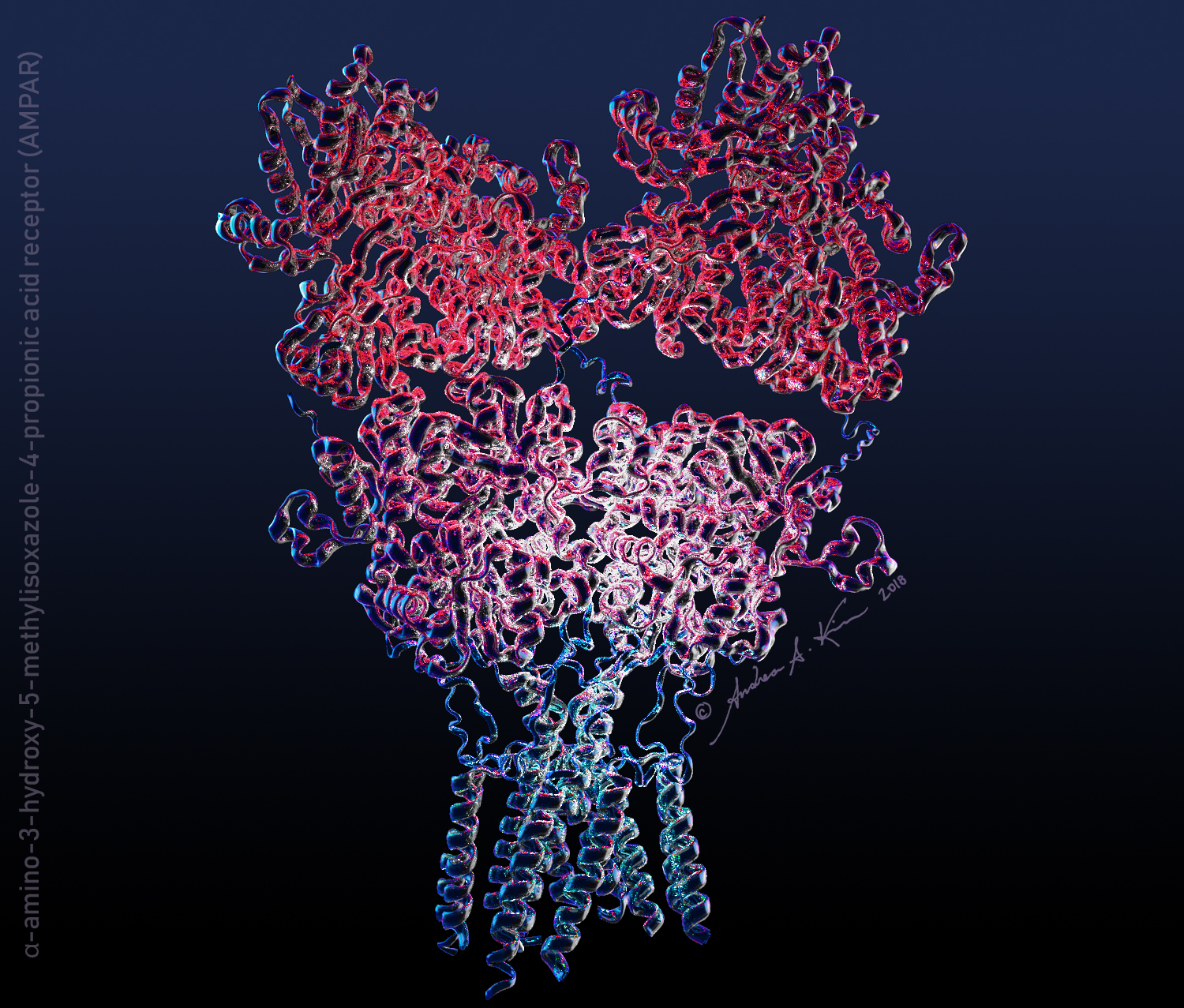
Everything starts with pencil on paper, and in this case, over 40 hours of scientific literature review! This piece was particularly difficult, as Uniprot (a database of protein sequence and functional information) and RCSB Protein Data Bank (a database for the 3D structural data of molecules) had mismatched mapping of the alpha-amino-3-hydroxy-5-methylisoxazole-4-propionic acid receptor (AMPAR - the biggest molecule in my scene!)
Identifying zeta-inhibitory peptide sequence
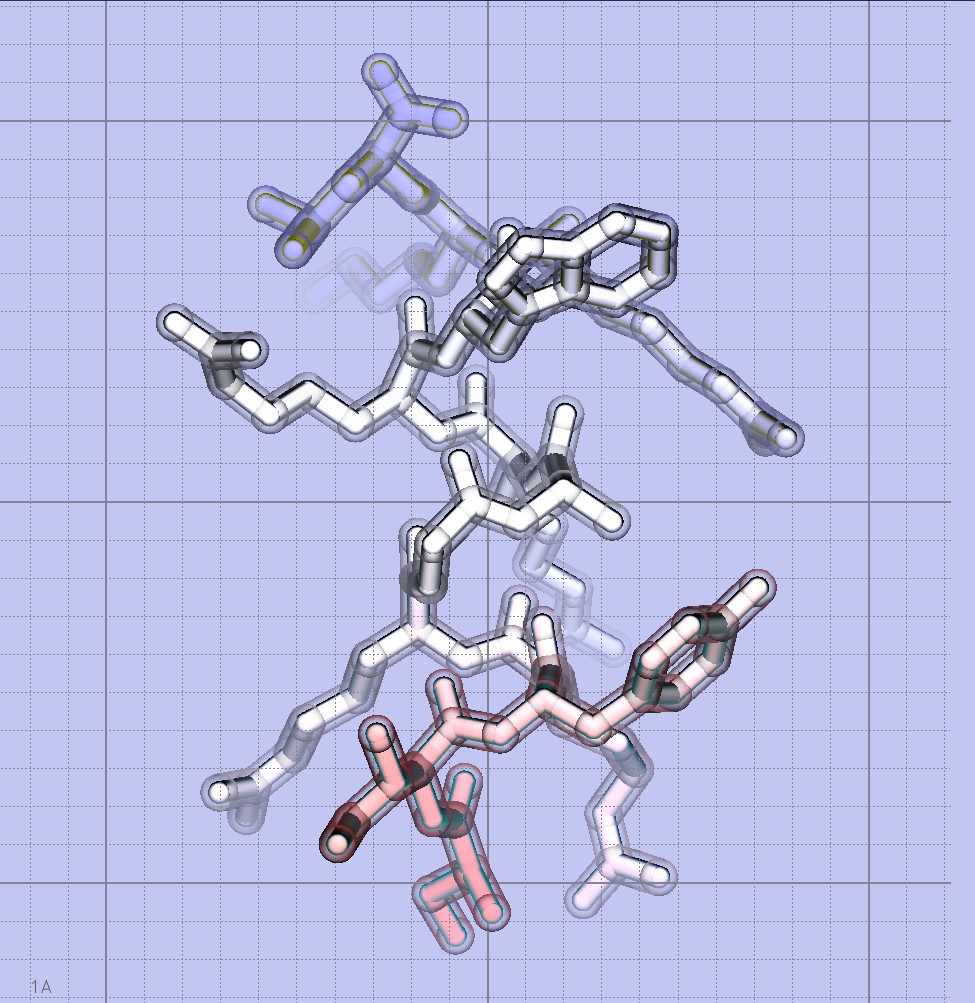
The 13 amino-acid pseudosubstrate sequence of zeta-inhibitory peptide (ZIP) was identified in literature and confirmed in Uniprot. Obtained from the regulatory domain of the full-length PKCz, I learned that ZIP actually has to be conjugated on its N-terminal to a myristoyl group in order to achieve peptide penetration through cells.
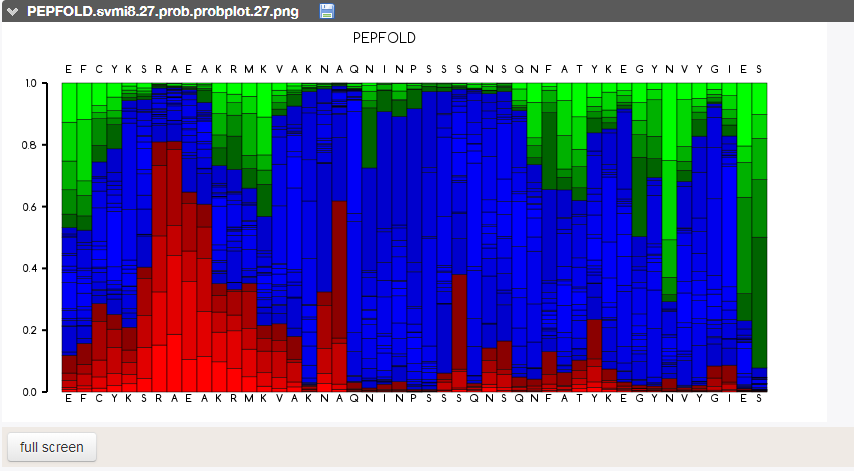
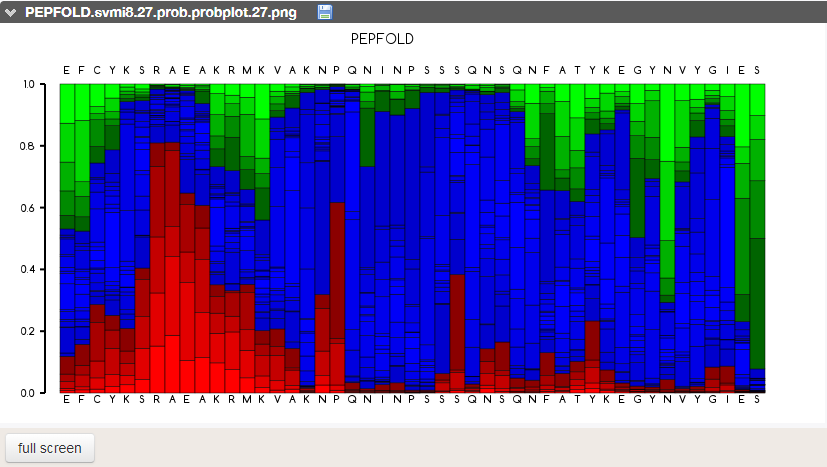
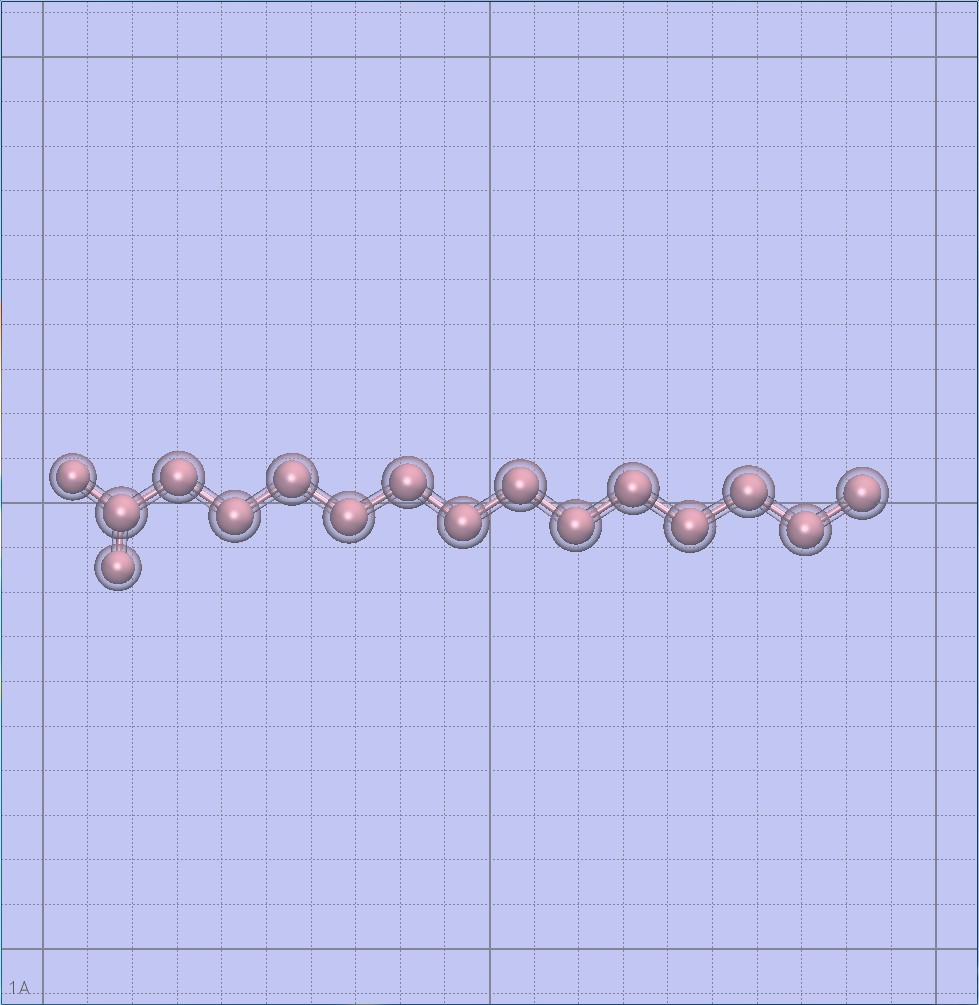
Prediction of peptide fold
As ZIP is an intrinsically disordered peptide (i.e. crystallization cannot be achieved with current technology), I had to insert the sequence into PEP-FOLD3, a de novo approach aimed at predicting peptide structures from amino acid sequences. This method couples the predicted series to an algorithm, returning the error report seen above. This looks pretty good!
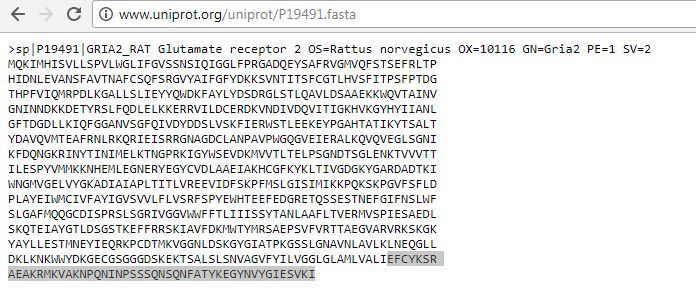
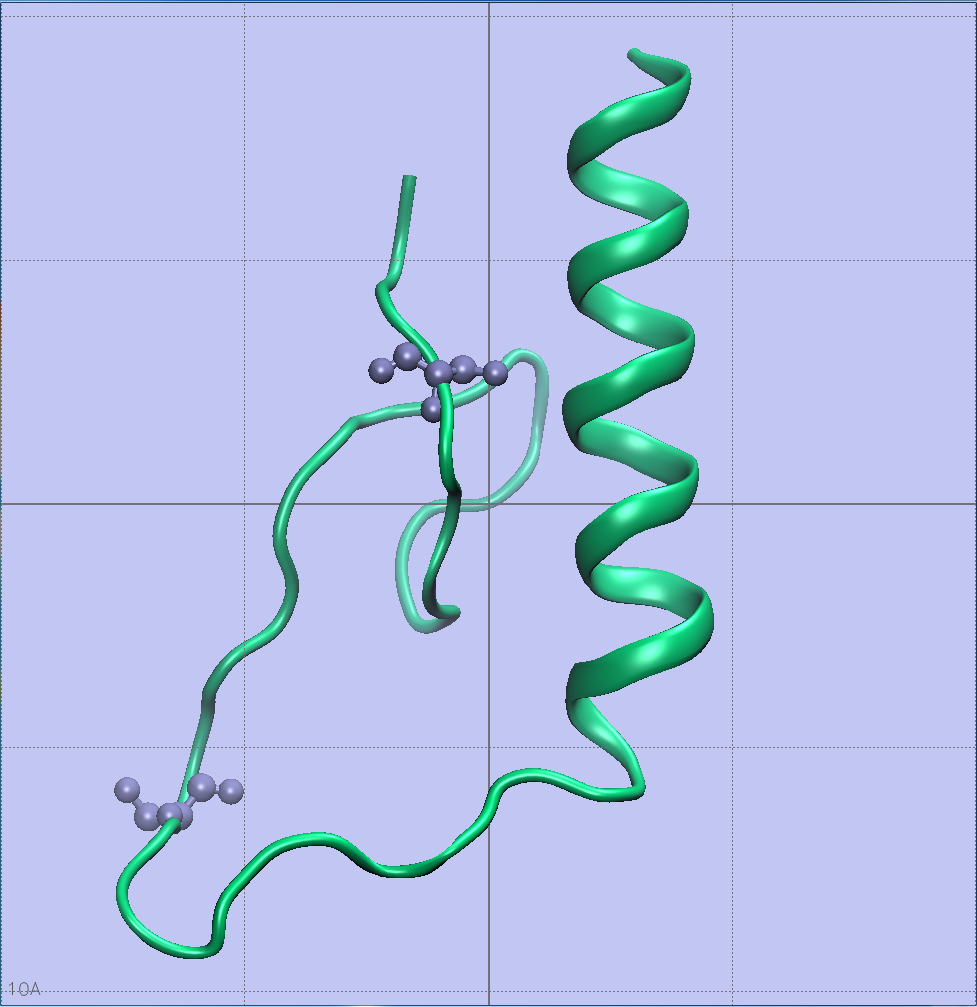
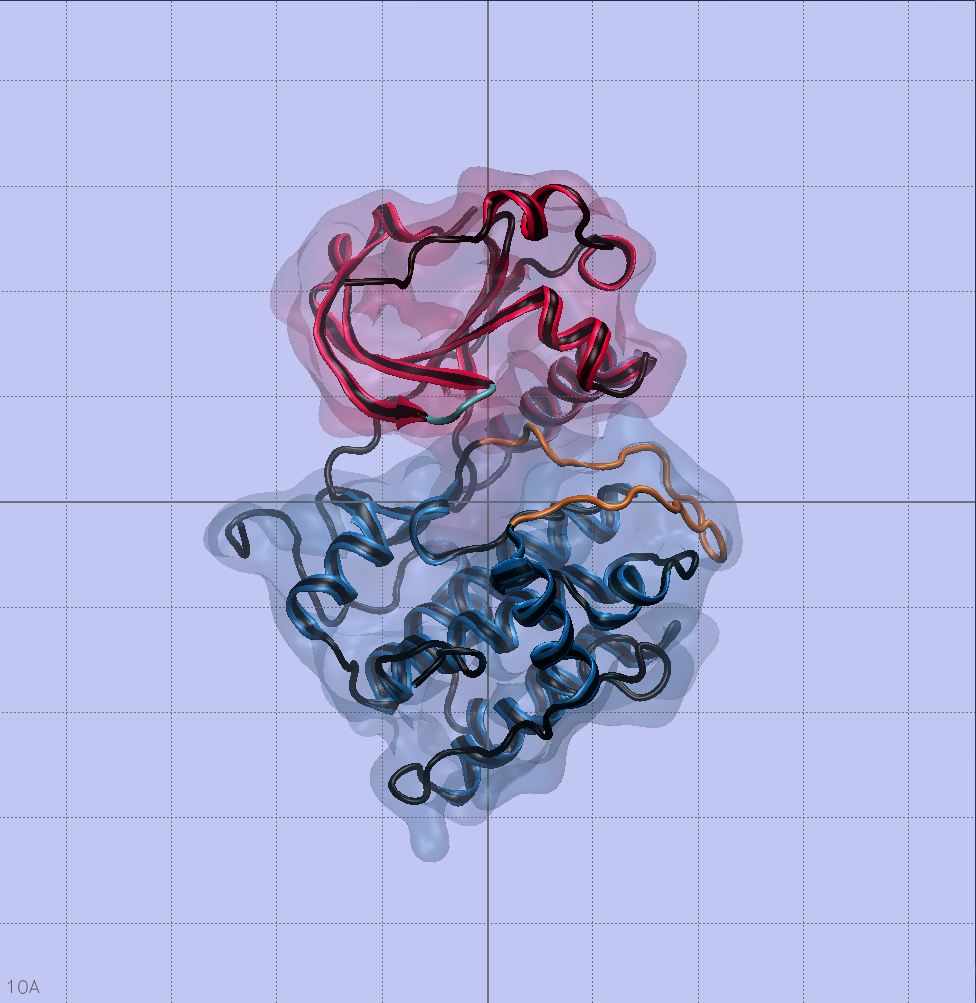
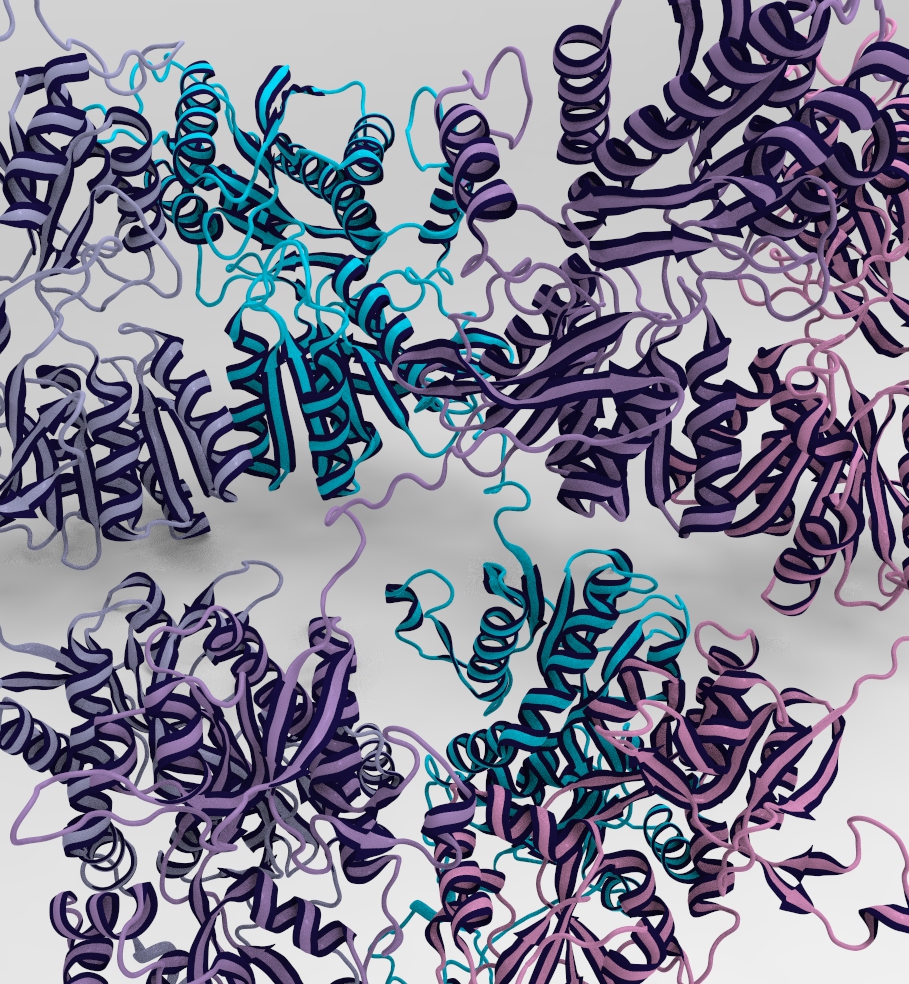
The Serine phosphorylation sites of AMPA GluR2 Receptor
Phosphorylation sites of AMPAR that my piece highlights are serines 863 and 880. Using the sequence viewer from Visual Molecular Dynamics, I was able to isolate the two amino acids of interest shown to the left. Yes, I went through every single amino acid to ensure I had the correct ones.
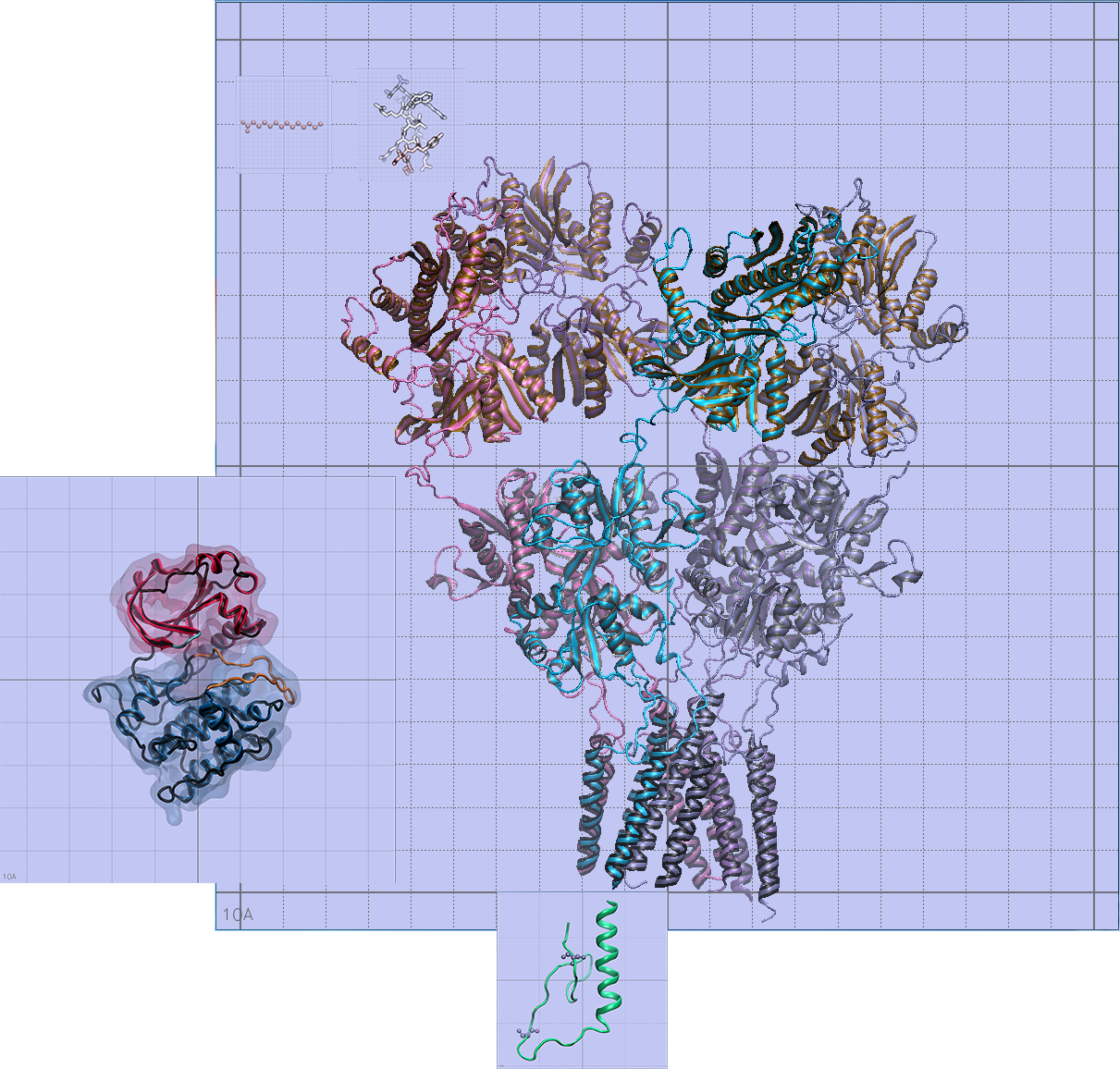
Confirming size relationships and orientation of the different molecules
I used built-in measurement tools in VMD to the nearest Angstrom to confirm size relationships between all of my models, and the orientation of the transmembrane helices in the lipid bilayer using OPM (Orientations of Proteins in Membranes).
This didn't take very long, but it's one of the most important steps to take in ensuring scientific accuracy.
Bonus images I rendered for fun!!
...because this project took an extensive amount of time, I made sure to have fun with it :)